Par Kristin Bartik
L’eau joue probablement un rôle indispensable pour l’apparition le la vie. Dans cet article, Kristin Bartik [1] nous décrit le rôle essentiel que joue ce composé dans la structuration des macromolécules biologiques, mais aussi leur stabilité et leur dynamique. L’eau est maintenant considérée comme partie intégrante des systèmes biomoléculaires.
It is clear that life on Earth depends on the presence of liquid water. In living systems water is pervasive and ubiquitous and cannot be considered as a simple dilutent. It performs many functions : it transports, structures, stabilizes, lubricates, reacts and partitions. Proof for the finely tuned involvement of water in life processes is provided by the fact that heavy water is toxic to these processes.
Proteins and nucleic acids play important biological functions : they catalyze and regulate reactions, transport substrates, code and transcribe genetic information. It is widely appreciated that water molecules play an invaluable role in governing the structure, stability, dynamic, and function of these biomolecules. Indeed, they lack activity in the absence of water. It is however only in recent years that water has been quantitatively treated as an integral component of biomolecular systems.
A variety of experimental and computer studies acknowledge the active role of water in biomolecule structure, stability and dynamics. Experimentally information is obtained from x-ray diffraction, neutron diffraction, nuclear magnetic resonance (NMR) and femtosecond spectroscopy measurements [1-3]. Molecular dynamics simulations can complement the knowledge gained through experiments and offer a description of the biomolecule and solvent as well as the time dependency of their dynamics[4 ;5].
The role of water in the structure, stabilty and dynamics of proteins and nucleic acids is discussed below. Its importance in molecular recognition processes that involve these biological macromolecules is also addressed.
I. Role of water in protein folding, structure and stability
Proteins are polymers of alpha-amino-acids which, under normal pH, temperature and ionic strength conditions, generally adopt a preferred three dimensional (3D) fold. The side chains of the 20 amino acids that compose proteins possess different functional groups (amide, carboxylic, hydroxyl, thiol, aromatic, …).
The sequence of amino-acids in the protein polymer is known as its primary structure. The distribution of the atoms in space is described in terms of secondary and tertiary structure which characterize, respectively, the elements of regular secondary structure (alpha-helices and beta-sheets, … ) and the global fold (alpha-barrels, 4 helix bundles, … ) of the protein. It is however interesting to point out that theoretical models predict that only a limited number of protein folds exist and that the discovery of an entirely new fold will be a rare event in the future [6 ;7].
The term primary structure misleadingly suggests that the connectivity in a biological macromolecule is its primary structural descriptor. The protein sequence alone, however, is insufficient to predict the protein 3D structure. Proteins with different sequences can have near identical 3D structures while small changes in a sequence can lead to different folds. In the case of prion-type proteins we are even confronted with the situation where a protein can adopt two completely different folds.
The interactions between non-covalently bound atoms (non-covalent interactions) play important roles in defining the structure and stability of native (biologically active) proteins. Solution thermodynamic and spectroscopic studies are performed specifically to try and understand and quantify these non-covalent interactions. The difference in free energy between native and unfolded states of a protein is due to enthalpic and entropic contributions which generally compensate each other. The stability margin of native proteins relative to their denatured inactive forms hardly exceeds 50 kJmol-1 [8]. This slender stability margin ensures that proteins maintain enough flexibility to fulfil their diverse roles. Many forces make small and conflicting contributions to protein stability. The current understanding of the way these forces “interact” is incomplete ; as often as not mutations (replacement of one amino acid by another) have the opposite effect to that which had been predicted. It is important to point out that while various experimental techniques can elucidate the structure of native proteins, a similar detailed description of inactive (denatured) states still elucidates present day investigators. The denatured/inactive state is certainly not “random coil” and completely flexible. The ignorance of the structural details of unfolded proteins makes it difficult to analyse all data quantitatively.
Electrostatic interactions, among which the so called “salt bridges” (coulombic interactions between opposite charges) and “cation-” interactions (charge-aromatic cycle), and van der Waals (London) interactions play important roles in defining the stability of proteins. London interactions are weak and non-directional but are important due to the fact that they are omni-present. The hydrophobic effect and hydrogen bonds are also of prime importance and water is an actor in these contributions to protein structure and stability [9]. They will therefore be discussed here in more detail. Figure 1 is a schematic representation of the way in which water is involved in protein structure and stability.
The hydrophobic effect is generally considered to be the major driving force for the folding of globular proteins. It results in the burial of the hydrophobic amino acid side-chains in the core of the protein. Based on the work of Frank and Evans [10], which explains at the molecular level the low solubility of non-polar species in water, Kauzmann introduced already in the late 1950’s the concept of ‘hydrophobicity’ to explain the complexities of protein folding [11]. Water tends to form ordered cages around non-polar groups (hydrophobic hydration) which leads to a decrease in entropy of the system. These water molecules gain entropy when they are released after hydrophobic surfaces are put in contact with each other. This contributes in a very favorable way to the free energy of stabilisation of the protein. Water is therefore fundamental in protein folding because of its role in defining hydrophobic attractions.
The hydrophobic “collapse” of the protein is necessarily accompanied, and guided, by hydrogen-bond formation between favorable functional groups [12]. A hydrogen bond (H-bond) occurs when two electronegative atoms, such as nitrogen and oxygen, interact with the same hydrogen. The strength of a hydrogen bond is around 2 to 20 kJ/mol. This is however not necessarily the amount of energy that the hydrogen bond contributes towards the stabilization of a folded protein. Indeed, in the unfolded state, potential hydrogen bonding partners in the polypeptide chain are satisfied by hydrogen bonds to water. When the protein folds these protein-to-water H bonds are broken, some are replaced by intra-protein H-bonds and the entropy of the solvent increases. The balance between the entropy and enthalpy terms is close but H-bonds make a positive contribution to protein stabilisation [13 ;14]. Despite the small contribu_ tion made to protein stability by hydrogen bonds, if an intramolecular hydrogen bond in a protein is broken or deleted without the possibility of forming a compensating H-bond to solvent, the protein will be destabilized and can loose its structure.
.
Figure 1 : Schematic representation of the various ways that water molecules are implicated in protein structure and stability.
.
X-ray and NMR show that well ordered water molecules are an integral part of all folded proteins and water molecules have been found to be conserved among homologous proteins. NMR studies of protein hydration in aqueous solution gives residence times ranging between 10-2 and 10-8 s for these interior water molecules [2]. Some of them contribute to the structure and stability of proteins by bridging, via hydrogen bonding, different functional groups present in the protein. These bridging water molecules must be considered part of the system and a protein should be defined as a polypeptide + water system. Hydrogen bonding networks, involving several water molecules which link distinct parts of a protein, have also been observed experimentally. Mutations can affect the number of structural water molecules within the protein core and disrupt essential interactions mediated by water H-bonding, resulting in destabilisation [15]. It is also thought that buried water molecules contribute in certain cases to the stabilisation of protein conformations simply by filling pockets in the protein structure (nature abhors void).
It is interesting to point out that significant improvement in protein structure prediction has been reported by adding a water knowledge-based potential to an established Hamiltonian for protein structure prediction [16]. These studies indicate that water not only induces protein folding and binding but also actively participates via water-mediated contacts. « Wet » Hamiltonians are expected to predict more accurate structures.
Water at the surface of biological macromolecules defines a layer, which has been termed « biological water » [17]. The hydration water in the proximity of the protein surface exhibits dynamical properties markedly deviating from those of bulk and residence times in the sub-nanosecond range. These water molecules play an important role in recognition processes which will be discussed in section 3.
II. Role of water on the structure and stability of nucleic acids
The basic repeating unit in deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) is the nucleotide [18]. It is composed of a cyclic sugar (deoxyribose in DNA or ribose in RNA) which is phosphorylated in one position and substituted by one of four different heterocyclic aromatic bases in another. The bases are adenine (A), guanine (G), cytosine (C) and thymine (T) in DNA and thymine is replaced by the functionally equivalent uracil (U) in RNA. The nucleotides are linked together via the phosphate groups (phosphodiesters bonds). The specific functions of DNA and RNA molecules are modulated by their 3D structure which depends not only on nucleotide sequence and base composition but also greatly on the environmental conditions.
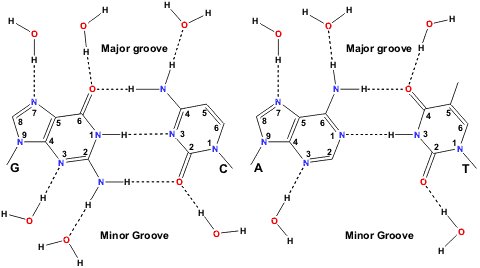
DNA is double stranded (the two strands are antiparallel to one another) and is usually thought of in terms of a right-handed double helix with complementary bases (C-G and A-T) pairing via H-bonds to each other (Figure 2). The base-pairs are located in the centre of the double helix, are coplanar and perpendicular to the helix axis. The outer envelope of the double helix is not cylindrically smooth but can display two grooves of different widths and depths. This three dimensional structure is capable of considerable variation and can undergo structural transitions between a number of different conformations. Whilst some of these changes are quite subtle, others are very dramatic and can, for example, involve a complete reversal of the handedness of the double helix. Five major structural variants of the DNA double helix have been identified. These have been designated A-DNA, B-DNA, C-DNA, D-DNA, Z-DNA (or S-DNA). Right-handed B-DNA is the predominant structural form in vivo and the two grooves are equally deep but do not have the same width. In right-handed A-DNA the two grooves do not have the same depth or width, the narrower one being deeper. Left handed Z-DNA is favoured at high salt concentrations and presents a single groove. RNA’s are not double stranded but adopt regular folds and usually more than half of the nucleotides participate in base-pairing. Consecutive stacking of the base-pairs gives rise to A-form type helices connected by single stranded stretches.
.
Figure 2 : Watson-Crick base-pairing and possible sites for water H-bonding to the paired bases. (from http://www.lsbu.ac.uk/water/nucleic.html)
.
Nucleic acids are highly charged polyanions. The neutralizing charges are provided by counterions in the surrounding aqueous medium. Metals like Mg++, Ca++, Na+ and K+ are present in milli-molar concentrations in the body and nucleic acids occur as complexes coordinated with these metals. Apart from this diffuse counterion atmosphere, specific sites for metal binding also exist [19]. Mg++ is for example known to play an important role in the stabilization of the tertiary structure of tRNA [20].
Along with hydrogen bonding between base-pairs and London dispersion forces between the stacked bases, water contributes to the stabilization of RNA and of DNA structures [20-23]. The role of water in the stabilization of the 3D structure of nucleic acids is even more important than in proteins because of the presence of negatively charged phosphate groups. Phosphate-phosphate electrostatic repulsion is diminished in water by the high dielectric constant of water. The degree of hydration of nucleic acids also plays a key role in their conformation. For example high water activity favors the B form of DNA and reduced water activity (or increased ionic strenght) leads to a transition from the B form to the C and A forms and if sequence permits to the D and Z-DNA forms.
Like in proteins, water is an integral part of nucleic acid structures. A shell of hydration, impermeable to cations, is located around DNA double helices [18]. It consists of about 18-19 water molecules per nucleotide in B-DNA and of about 13-14 in A-DNA. These water molecules are specifically bound to the phosphate groups and to the bases. X-ray studies show that there are six hydration sites per phosphate and that the positions and occupancies of these sites are dependent on the conformation and type of nucleotide [22-24]. Studies on A-DNA crystal structures have shown that water molecules bridge neighbouring phosphate groups and thus stabilize the A-form at lower water activities. The water molecules around the phosphate groups are not permanently situated however, due to the rather diffuse electron distribution of the phosphate groups. The paired bases are capable of hydrogen-bonding to water within the grooves (Figure 2). A spine of hydration is for example observed by x-ray crystallography in the minor groove of B-DNA. Energy calculations suggest that the presence of this spine of hydration is the prime reason for the narrowing of the minor groove. Hydration more persistent around the bases than around the phosphate groups but the residence times that are reported for these water molecules are still relatively short compared to those of bound water molecules in proteins. A second hydration shell, indistinguishable from bulk water as far as permeability to ions is concerned, surrounds the DNA double helix. In this shell the properties of the water molecules are different from those of bulk water even if residence times are in the sub-nanosecond range. Like for proteins this shell of hydration plays an important role in recognition processes which will be discussed in the following section.
RNA has a greater extent of hydration than DNA due to its extra oxygen atoms (the ribose O2’) and unpaired base sites. Stable hydration patterns are observed, like in DNA around double stranded regions [20 ;25].
III. Role of water in molecular recognition involving bio-molecules
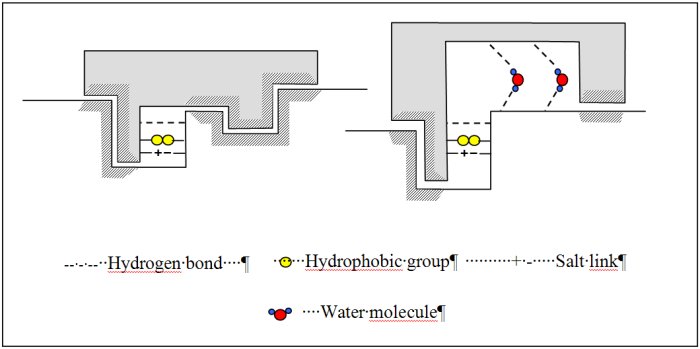
Binding processes are ubiquitous in biological systems ranging from the binding of small molecules like drugs to binding of proteins to DNA. Most, but not all, of these binding processes are highly specific with respect to the selection of binding sites [26-28]. Molecular recognition studies focused for a long time on the geometrical complementarity between the partners, eventually after conformational changes (Figure 3). This assumes that the driving forces for binding originates from direct interaction between the partners (attractive London interactions and presence of matching functional groups leading to H-bonding, salt bridges). Binding processes however generally occur in water. The fact that water molecules are abundantly observed experimentally at the interface between (bio)molecules suggests that water is indispensable for biomolecular recognition and self-assembly.
Water is a highly versatile component at the interface of biomolecular complexes. It can act both as a hydrogen bond donor and acceptor, imposing few steric constraints on bond formation and can take part in multiple hydrogen bonds. Water can thus confer a high level of adaptability to a surface allowing promiscuous binding yet it can also provide specificity and increased affinity to an interaction. Water in protein-ligand interactions can for example function as an extension of protein structure allowing varied ligands to be accommodated in a given binding site [28] or increasing affinity for specific ligand (Figure 3). For these water molecules, the energetic gain from water-mediated contacts is greater than the entropic cost resulting from their immobilization. Inclusion of water molecules in structure based ligand design is however currently largely overlooked because the structural and thermodynamic effects of water’s inclusion are hard to determine and model. (Bio)molecular associations, especially protein-protein and protein-DNA complexes, will of course also be stabilized as a result of the hydrophobic effect and of the screening, by water, of charge repulsion.
.
Figure 3 : Schematic representation of various models explaining protein-ligand interactions.
.
IV. Conclusion
Both hydrophobic and hydrophilic effects are dominant driving forces for biochemical processes : protein folding, nucleic acid stability and molecular recognition/binding events. Water, without any doubt, must be considered an integral part of biological macromolecules. The living world should be thought of as an equal partnership between proteins, nucleic acids and water.
V. References
- 1 Meyer, E. Internal water molecules and H-bonding in biological macromolecules : a review of structural features with functional implications ; Protein science 1, 1543-62 (1992)
- 2 Otting, G. NMR studies of water bound to biological molecules ; Progress in Nuclear Magnetic Resonance Spectroscopy 31, 259-285 (1997)
- 3 Pal, S. K., Peon, J., Bagchi, B. and Zewail, A. H. Biological Water : Femtosecond Dynamics of Macromolecular Hydration Journal of Physical Chemistry B 106, 12376-12395 (2002)
- 4 Feig, M. and Brooks, C. L. Recent advances in the development and application of implicit solvent models in biomolecule simulations Current Opinion in Structural Biology 14, 217-224 (2004)
- 5 Beveridge, D. L. and McConnell, K. J. Nucleic acids : theory and computer simulation, Y2K Current Opinion in Structural Biology 10, 182-196 (2000)
- 6 Govindarajan, S., Recabarren, R. and Goldstein, R. A. Estimating the total number of protein folds Proteins : Structure, Function, and Genetics 35, 408-414 (1999)
- 7 Mittl, P. R. E. and Grutter, M. G. Structural genomics : opportunities and challenges Current Opinion in Chemical Biology 5, 402-408 (2001)
- 8 Franks, F. Protein stability : the value of ’old literature’ Biophysical Chemistry 96, 117-127 (2002)
- 9 Levy, Y. and Onuchic, J. N. Water and proteins : a love- hate relationship Proceedings of the National Academy of Sciences of the United States of America 101, 3325-3326 (2004)
- 10 Franks, H. S. and Evans, M. W. Free volume and entropy in condensed systems III. Entropy in binary liquid mixtures. J. Chem. Phys. 13, 507-532 (1945)
- 11 Kauzmann, W. Some factors in the interpretation of protein denaturation Advances in Protein Chem. (C. B. Anfinsen, M. L. Anson, Kenneth Bailey, and John T. Edsall, editors, Academic Press Inc.) 14, 1-63 (1959)
- 12 Fernandez, A., Kardos, J. and Goto, Y. Protein folding : could hydrophobic collapse be coupled with hydrogen-bond formation ? FEBS Letters 536, 187-192 (2003)
- 13 McDonald, I. K. and Thornton, J. M. Satisfying hydrogen bonding potential in proteins Journal of Molecular Biology 238, 777-93 (1994)
- 14 Pace, C. N., Shirley, B. A., McNutt, M. and Gajiwala, K. Forces contributing to the conformational stability of proteins FASEB Journal 10, 75-83 (1996)
- 15 Covalt, J. C., Jr., Roy, M. and Jennings, P. A. Core and Surface Mutations Affect Folding Kinetics, Stability and Cooperativity in IL-1b : Does Alteration in Buried Water Play a Role ? Journal of Molecular Biology 307, 657-669 (2001)
- 16 Papoian, G. A., Ulander, J., Eastwood, M. P., Luthey-Schulten, Z. and Wolynes, P. G. Water in protein structure prediction Proceedings of the National Academy of Sciences of the United States of America 101, 3352-7 (2004)
- 17 Pal, S. K. and Zewail, A. H. Dynamics of Water in Biological Recognition Chemical Reviews (Washington, DC, United States) 104, 2099-2123 (2004)
- 18 Saenger, W. Principles of Nucleic Acid Structure (1984) Springer-Verlag ed.
- 19 Denisov, V. P. and Halle, B. Sequence-specific binding of counterions to B-DNA Proceedings of the National Academy of Sciences of the United States of America 97, 629-633 (2000)
- 20 Hermann, T. and Patel, D. J. Stitching together RNA tertiary architectures Journal of molecular biology 294, 829-49 (1999)
- 21 McConnell, K. J. and Beveridge, D. L. DNA structure : what’s in charge ? Journal of molecular biology 304, 803-20 (2000)
- 22 Berman, H. M. Crystal studies of B-DNA : the answers and the questions Biopolymers 44, 23-44 (1997)
- 23 Wahl, M. C. and Sundaralingam, M. Crystal structures of A-DNA duplexes Biopolymers 44, 45-63 (1997)
- 24 Schneider, B., Patel, K. and Berman, H. M. Hydration of the phosphate group in double-helical DNA Biophysical Journal 75, 2422-2434 (1998)
- 25 Auffinger, P. and Westhof, E. RNA hydration : three nanoseconds of multiple molecular dynamics simulations of the solvated tRNAAsp anticodon hairpin Journal of Molecular Biology 269, 326-341 (1997)
- 26 Ben-Naim, A. Molecular recognition – viewed through the eyes of the solvent Biophysical Chemistry 101-102, 309-319 (2002)
- 27 Ladbury, J. E. Just add water ! The effect of water on the specificity of protein-ligand binding sites and its potential application to drug design Chemistry & Biology 3, 973-980 (1996)
- 28 Tame, J. R. H., Sleigh, S. H., Wilkinson, A. J. and Ladbury, J. E. The role of water in sequence-independent ligand binding by an oligopeptide transporter protein Nature Structural Biology 3, 998-1001 (1996)
[1] Professeur au laboratoire d’ingénierie moléculaire et biomoléculaire, à l’Université Libre de Bruxelle.






Bonjour, un de mes amis travaille sur une thèse universitaire sur ce sujet et il a besoin d’aide avec un scénario complexe. Seriez-vous en mesure d’aider plz?
Bonjour,
Je vous suggère de lui conseiller de contacter directement l’auteur de l’article : Kristin Bartik.